DNA microscopy creates 3D imaging of entire organisms from inside out
University of Chicago researchers have developed a groundbreaking imaging technology that visualizes the spatial organization of genetic material in three dimensions, without relying on conventional optical instruments. This technique, called volumetric DNA microscopy, works by tagging DNA and RNA molecules and allowing neighboring tags to interact, creating a molecular network that encodes their relative positions and produces a comprehensive spatial map of gene expression down to individual cells.
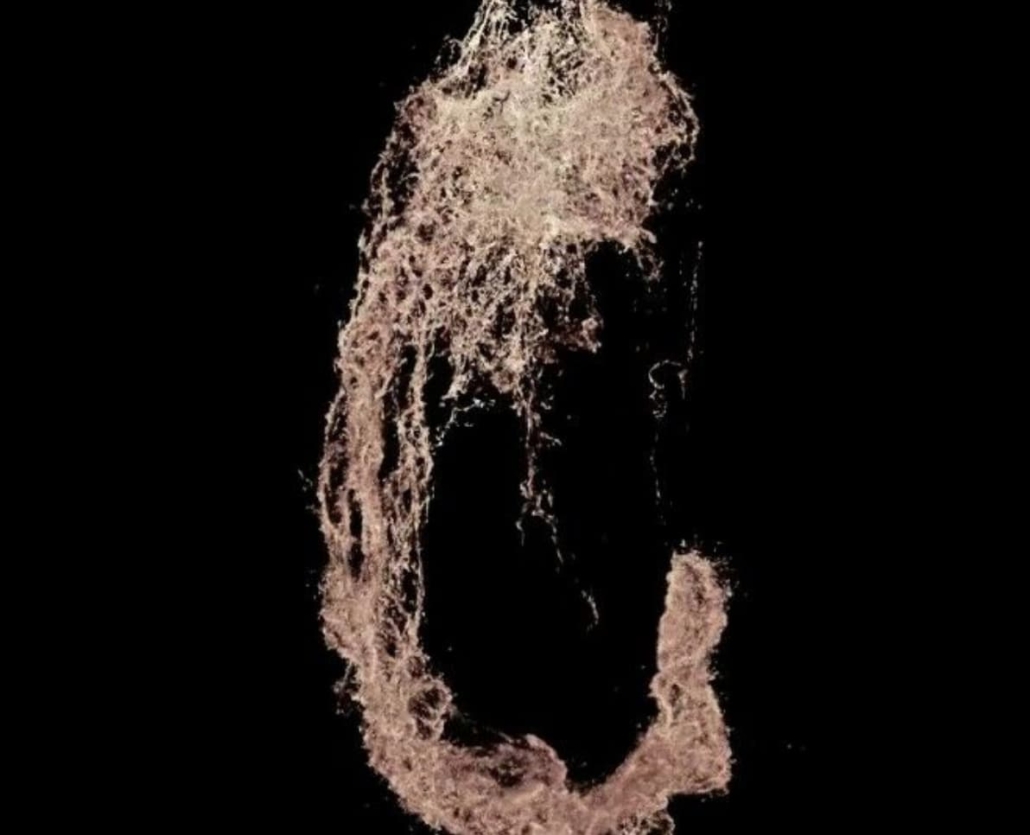
DNA microscopy image of a zebrafish embryo. © Weinstein Lab
The inside-out imaging approach
Traditional microscopes use light or lenses to visualize specimens from the outside. DNA microscopy, however, takes a fundamentally different approach.
“It’s a level of biology that no one has ever seen before,” said Joshua Weinstein, PhD, Assistant Professor of Medicine and Molecular Engineering at UChicago and senior author of the study published in Nature Biotechnology. “To be able to see that kind of a view of nature from within a specimen is exhilarating.”
Rather than relying on external observation, DNA microscopy creates images by calculating molecular interactions within the sample itself. The technique initially tags DNA and RNA molecules with unique molecular identifiers (UMIs) – short DNA sequence tags that attach to genetic material throughout an organism.
These UMIs then begin making copies of themselves, which can interact with nearby UMIs to create unique event identifiers (UEIs). The frequency of these interactions directly correlates with physical proximity – molecules that are close to each other generate more UEIs than those farther apart.
“This turns the idea of imaging on its head,” Weinstein explained. “Rather than relying on an optical apparatus to shine light in, we can use biochemistry and DNA to form a massive network between molecules and encode their proximities to each other.”
Advanced computational reconstruction
After genetic sequencing, the researchers employed a novel computational approach called Geodesic Spectral Embeddings (GSE) to reconstruct the three-dimensional positions of each molecular tag.
This method overcomes previous limitations in processing the enormous volumes of data generated from the approximately 10 million molecules tagged throughout the sample. The researchers demonstrated the technique’s effectiveness by creating complete DNA images of zebrafish embryos, common model organisms for studying development and neurobiology.
The computational innovation is particularly significant because it allows researchers to handle the complexity of three-dimensional biological structures across multiple length scales. By combining both short-range and long-range molecular interactions into a single dataset, the technique can capture the intricate details of biological structures at different resolutions simultaneously.
Applications in complex tissues
The researchers believe DNA microscopy could be particularly valuable for understanding genetic expression in complex tissue environments that create numerous unique mutations, such as tumors.
“This is the critical foundation for being able to have truly comprehensive information about the ensemble of unique cells within the lymphatic system or tumor tissue,” Weinstein said. “There has still been this major gap in technology for allowing us to understand idiosyncratic tissue, and that’s what we’re trying to fill in here.”
The technique doesn’t require prior knowledge of the genome or shape of a specimen, making it suitable for analyzing genetic expression in unique biological contexts. It could potentially help unravel complex immune system interactions and guide more precise treatments.
In the paper, the authors note that “our extension of spatial genetic measurements to three dimensions, independent of prior templates, opens the door to detailed joint resolution of genomics and morphology in biological tissues.” This capability could significantly advance our understanding of how genetic information translates into physical structures in living organisms.
Layered chemical approach
To enable three-dimensional imaging at different scales, the researchers developed a sophisticated multi-step chemical process. First, RNA molecules in fixed cells are reverse-transcribed to complementary DNA and tagged with UMIs. These UMIs are then amplified through a process called rolling circle amplification (RCA), creating DNA “nanoballs” that remain anchored near their points of origin.
This anchored amplification ensures that interactions between UMIs reflect their true spatial relationships. The researchers then introduced a controlled diffusion process through in vitro transcription inside a reversible polyethylene glycol hydrogel, which allowed for capturing longer-range interactions between molecules.
By combining these approaches, the technique encodes both local and global spatial relationships into the final dataset, enabling comprehensive mapping of genetic expression throughout the entire organism.
The work represents a significant leap forward in spatial genomics, offering researchers a new window into the molecular organization of complex biological systems from the inside out.
Reference
Qian, N., & Weinstein, J. A. (2025). Spatial transcriptomic imaging of an intact organism using volumetric DNA microscopy. Nature Biotechnology.
https://doi.org/10.1038/s41587-025-02613-z

