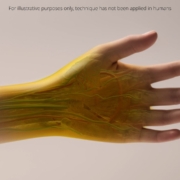
Groundbreaking technique renders skin transparent, opening new avenues for diagnostic imaging
Researchers have developed a novel method to make skin and underlying tissues transparent using a common food dye, heralding potentially far-reaching applications for a wide range of medical diagnostics and certain treatments.


 BiVACOR
BiVACOR

 WHO
WHO