AI system designs peptides to target previously ‘undruggable’ proteins
Biomedical engineers at Duke University have developed an artificial intelligence platform that can design short proteins capable of binding to and destroying previously untreatable disease-causing proteins. The breakthrough could revolutionise drug development for currently intractable diseases.
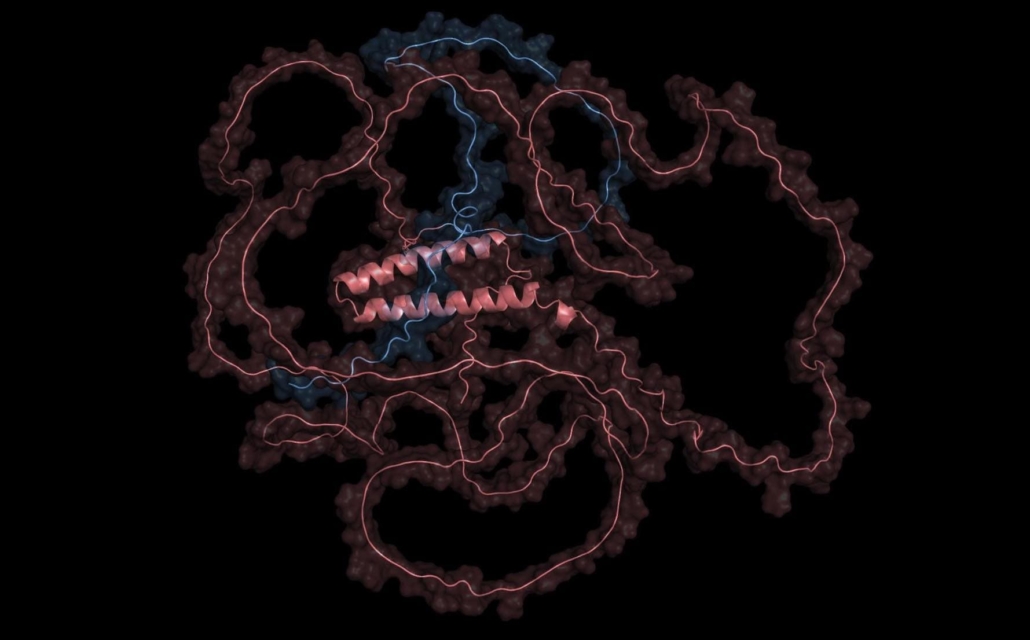
The image shows one of the tangled, unstructured proteins Chatterjee and his team are hoping to find binders for. © Pranam Chatterjee, Chatterjee Lab, Duke University
The innovative system, called PepPrCLIP (Peptide Prioritization via CLIP), takes inspiration from OpenAI’s image recognition technology. Instead of matching images to text descriptions, the system matches newly designed peptides to specific disease-causing proteins.
“OpenAI’s CLIP algorithm connects language with an image. If you have text that says ‘dog,’ you should get an image of a dog,” explains Pranam Chatterjee, assistant professor of biomedical engineering at Duke. “Instead of language and image, we trained it to match peptides and proteins. PepPr makes the peptides, and our adapted CLIP algorithm will screen those peptides and tell us which ones will make a good match.”
Tackling the undruggable challenge
More than 80% of disease-causing proteins have been considered “undruggable” because their complex, disordered structures make them difficult for conventional drugs to target effectively. While existing therapies work well with proteins that have well-defined structures, they struggle with proteins that resemble “a messy ball of yarn.”
The research team demonstrated PepPrCLIP’s capabilities through several increasingly challenging tests. The system successfully designed peptides that could bind to and inhibit both simple enzyme proteins and more complex cancer-related proteins.
Breakthrough in cancer protein targeting
In their most ambitious test, the team created peptides targeting a highly disordered protein associated with synovial sarcoma, an aggressive cancer affecting young people. According to Chatterjee, “It’s like a bowl of spaghetti. It’s the most disordered protein in the world.”
The researchers tested ten designed peptides in synovial sarcoma cells and found they could successfully bind to and degrade the target protein. This achievement demonstrates the potential for developing treatments for previously untreatable cancers.
Future therapeutic applications
In their paper’s discussion, the authors note that their “two-step method represents a first protein binder design module from the target sequence alone without any requirement of 3D structural information in the generation process.” This breakthrough opens new possibilities for treating various diseases caused by disordered proteins.
The team plans to partner with medical and industry professionals to develop peptides for therapeutic applications, including treatments for Alexander’s Disease, a fatal neurological condition primarily affecting children, and various types of cancer.
“These complex, disordered proteins have made a lot of cancers and diseases practically undruggable because we couldn’t design molecules that bind to them,” said Chatterjee. “But PepPrCLIP showed that it could work on even the most complicated protein, and that opens up a lot of exciting clinical possibilities.”
Reference:
Bhat, S., Palepu, K., Hong, L., et. al. (22 Jan 2025). De novo design of peptide binders to conformationally diverse targets with contrastive language modeling. Science Advances, Vol 11, Issue 4. doi: https://doi.org/10.1126/sciadv.adr8638.

