New NextBrain atlas enables unprecedented granularity in human brain imaging analysis
Researchers at University College London have developed NextBrain, a probabilistic histological atlas of the entire human brain that enables automated segmentation of magnetic resonance imaging scans into 333 distinct regions of interest. Built from approximately 10,000 histological sections across five brain hemispheres, the atlas combines artificial intelligence-enabled registration techniques with Bayesian segmentation methods to provide unprecedented anatomical detail for neuroimaging studies.
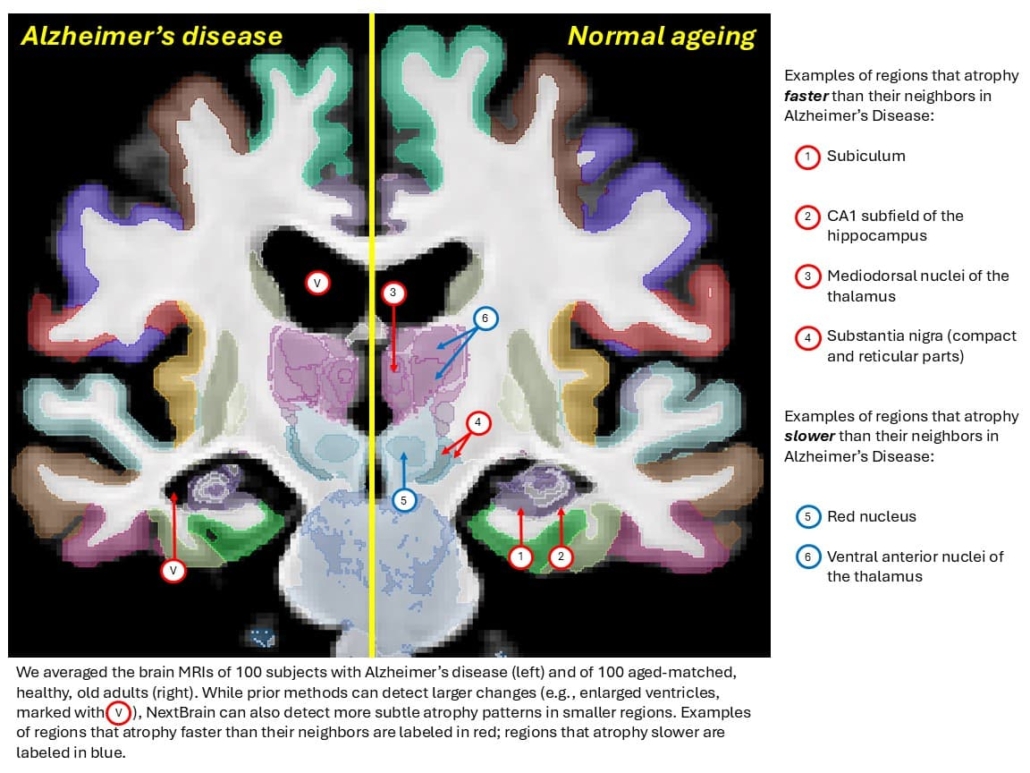
© Martina Bocchetta, Dorit Kliemann and Juan Eugenio Iglesias
Advanced registration framework achieves submillimetre accuracy
The NextBrain project addresses a fundamental limitation in neuroimaging: existing histological atlases either lack probabilistic labels throughout the whole brain or provide insufficient anatomical granularity for detailed regional analysis. The research team, led by Juan Eugenio Iglesias at UCL’s Department of Medical Physics and Biomedical Engineering, developed a sophisticated computational pipeline to reconstruct three-dimensional volumes from serial histology sections stained with haematoxylin and eosin and Luxol fast blue.
The registration framework incorporates three novel components specifically designed for this challenge. A differentiable regulariser minimises overlap between tissue blocks whilst eliminating gaps in the reconstruction. An artificial intelligence registration method employs contrastive learning to achieve accurate alignment between MRI and histology modalities. Finally, a Bayesian refinement technique based on Lie algebra ensures three-dimensional smoothness across the reconstruction, even when tissue folding or tearing creates outliers.
Quantitative evaluation using 250 manually placed landmark pairs demonstrated a mean registration error of 0.99 millimetres with a standard deviation of 0.51 millimetres. This represents a 32% reduction compared with the team’s previous pipeline, with statistical significance below P < 10⁻²¹ across all landmarks. “Our method yields an average error of 0.99 mm, which is a considerable reduction with respect to previous approaches,” the authors note in their discussion.
Multimodal dataset provides comprehensive neuroanatomical resource
Each of the five cases comprises accurately aligned high-resolution ex vivo MRI at 400-micrometre isotropic resolution, serial histology digitised at 4-micrometre resolution with 250-micrometre or 500-micrometre spacing, and region-of-interest segmentations obtained through semi-automated artificial intelligence methods. The anatomical protocol includes 34 cortical labels following the Desikan-Killiany atlas and 299 subcortical labels derived from various atlases for different brain regions.
The probabilistic atlas was constructed by coregistering labels from the five hemispheres using diffeomorphic registration methods that preserve neuroanatomical topology. Crucially, the team initialised the atlas building process with the Montreal Neurological Institute template, which serves to prevent biases towards any individual case whilst anchoring the atlas to a well-established common coordinate frame derived from 305 subjects.
The resulting atlas models voxel-wide probabilities for 333 regions of interest on a 0.2-millimetre isotropic grid, representing a substantial advance over existing whole-brain probabilistic atlases. The widely used SAMSEG atlas in FreeSurfer, for comparison, models only 13 brain regions at 1-millimetre resolution.
Validation demonstrates clinical utility across multiple applications
The research team validated NextBrain through multiple experimental paradigms. For ultra-high-resolution ex vivo MRI segmentation, they created a gold standard segmentation of a publicly available 100-micrometre isotropic whole brain scan, requiring over 100 hours of manual tracing effort despite using semi-automated techniques. Comparison yielded a median Dice overlap score of 0.667, comparable to other Bayesian segmentation methods for brain subregions.
In an Alzheimer’s disease classification task using 383 subjects from the Alzheimer’s Disease Neuroimaging Initiative, NextBrain achieved an area under the receiver operating characteristic curve of 0.953 with 90.3% classification accuracy. “The increased segmentation accuracy and granularity of NextBrain enables it to achieve AUROC = 0.953 and 90.3% accuracy—with a significant increase in AUROC with respect to the Allen MNI template (P = 0.01 for a DeLong test),” the authors report.
Fine-grained ageing analysis reveals regional specificity
Application to 705 subjects from the Ageing Human Connectome Project dataset revealed highly detailed patterns of brain ageing. The analysis demonstrated stronger negative correlations between age and volume in the anterior caudate compared with posterior caudate, and in the external segment of the globus pallidus compared with the internal segment. Within the thalamus, correlations proved most significant in the mediodorsal, anteroventral, and pulvinar subnuclei, which represent key regions in limbic, lateral orbitofrontal, and dorsolateral prefrontal circuits.
Validation using the OpenBHB meta-dataset comprising over 3,000 healthy individuals demonstrated that NextBrain consistently outperformed the Allen MNI template whilst producing segmentations with accuracy throughout the lifespan. Notably, NextBrain showed minimal age-related performance correlation (r = 0.046, P = 0.009) compared with the Allen MNI template’s strong negative correlation (r = −0.274, P < 10⁻⁵⁵).
The research team has publicly released all components of NextBrain, including raw and aligned data, an online visualisation tool, the probabilistic atlas, the Bayesian segmentation tool distributed with FreeSurfer, and ground truth delineations for the high-resolution ex vivo hemisphere used in validation. “By enabling researchers worldwide to automatically analyse brain MRIs at a higher level of granularity, NextBrain holds promise to increase the specificity of findings and accelerate our quest to understand the human brain in health and disease,” the authors conclude.
Reference
Casamitjana, A., Mancini, M., Robinson, E., et. al. (2025). A probabilistic histological atlas of the human brain for MRI segmentation. Nature. https://doi.org/10.1038/s41586-025-09708-2

